 MONTREAL, CANADA—When taking a dip this summer you will probably swallow tens, possibly hundreds, of microscopic plankton called choanoflagellates. These common organisms have led to an uncommon insight into how multicellular organisms might have evolved. Bacteria can prompt single-celled choanoflagellates to divide into multicellular versions of themselves, University of California (UC), Berkeley, biologist Nicole King reported last week here at the 71st annual meeting of the Society for Developmental Biology. King hopes the work will prompt biologists to look more closely at the role of microorganisms in the evolution of multicellularity.
MONTREAL, CANADA—When taking a dip this summer you will probably swallow tens, possibly hundreds, of microscopic plankton called choanoflagellates. These common organisms have led to an uncommon insight into how multicellular organisms might have evolved. Bacteria can prompt single-celled choanoflagellates to divide into multicellular versions of themselves, University of California (UC), Berkeley, biologist Nicole King reported last week here at the 71st annual meeting of the Society for Developmental Biology. King hopes the work will prompt biologists to look more closely at the role of microorganisms in the evolution of multicellularity.
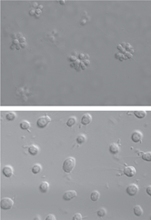
To the untrained eye, choanoflagellates look like animals. But they are less complex—the closest living relatives of animals but on an older branch of the tree of life. As such, these organisms can provide clues about what early animals looked like and can help reconstruct the events from more than 600 million years ago that led to the incredible diversity of the animal kingdom.
To investigate the transition to colony life, King decided to sequence the genome of a colony-forming choanoflagellate and compare it with the genome of a unicellular individual. But before sequencing, she asked undergraduate Richard Zuzow to purge the sample of everything but the plankton itself. When Zuzow added antibiotics to get rid of any bacteria, the choanoflagellate colonies disappeared. At first, “I didn’t believe him,” King recalls. But with repeated tests, she became convinced that “the bacteria are the important part of the [multicellular] story,” she says
:: Read more here (behind pay-wall) or download pdf here ::

